🗂️ Segmentation and Tracking Pipeline#
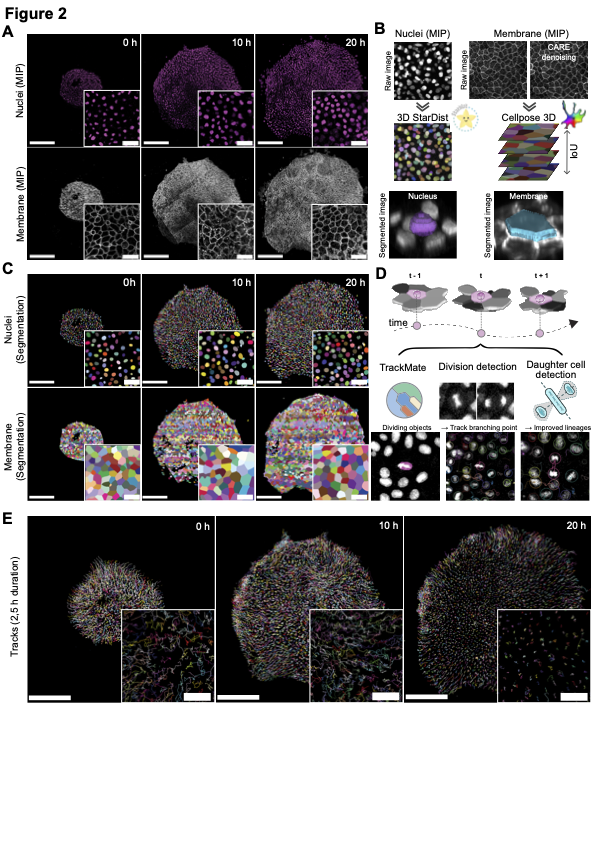
Confocal time-lapse stacks were first deskewed and stabilised: tiled volumes were stitched in ImageJ, then tilt artefacts were removed with a custom macro and residual sample drift was corrected frame-by-frame with Fast4DReg. For the membrane channel, photon-shot noise was reduced in three dimensions by a content-aware restoration network (CARE, CSBDeep), ensuring uniform signal-to-noise across developmental stages. To start the pipeline merge the nuclei and membrane channel and call it Merged.tif that you can put in a folder such as Mari_Second_Dataset_Analysis. Then update the config files with your data paths, model paths, the parameters specified here can be modifed but are set to the ones used for our analysis. Then you can run the zeroth step of the workflow that creates nuclei and memrane channel tif files.
⚛️ Nuclear masks#
Nuclei were segmented volumetrically with a bespoke StarDist 3-D model trained on manually annotated sub-volumes that sampled early, intermediate, and late time points. Training patches were augmented by random flips, rotations, and intensity perturbations to maximise robustness. Prediction was wrapped in the VollSeg pipeline: a UNet first identified the tissue region on Z-maximum projections, intensities were locally normalised, and only the cropped region was forwarded to StarDist. Batched GPU inference yielded integer-labelled nuclear volumes with high fidelity; evaluation against ground-truth volumes produced F1-scores > 0.9 and mean Object-at-Ground-Truth-Centroid Jaccard indices above 0.8. To run this step on the Raw nuclei stack generated in the zeroth step run the nuclei segmentation script
🧬 Membrane masks#
Prior to segmentation, membrane-channel stacks were first denoised with CARE (Content-Aware Image Restoration, CSBDeep) to suppress photon shot noise and improve edge contrast. Membrane segmentation then exploited the higher axial resolution of 2-D slices: a Cellpose 2.0 model—fine-tuned via the human-in-the-loop workflow starting from the “cyto2” backbone—was trained on hand-drawn contours from representative z-planes and time points. During inference each slice was segmented independently (flow_threshold = 1.0, cellprob_threshold = -1.0, diameter ≈ 33 px). To reconstitute full cell volumes, adjacent slice masks were stitched together by maximal pair-wise intersection-over-union (IoU ≥ 0.6); segments thinner than two planes were discarded. The resulting 3-D membrane labels, generated from denoised input, captured the apico-basal envelope with moderate mean IoU yet sufficient surface fidelity for downstream morphometrics. Denoising script followed by Membrane segmentation.
🔗 Integration and lineage reconstruction#
Membrane and nuclear objects were linked by nearest-neighbour matching of centroids in 3-D space, ensuring every cell acquired both a shape envelope and a nuclear anchor. Nuclear centroids then served as the basis for tracking in TrackMate 7: integer-label images were imported with the Label Image Detector, and trajectories were assembled using the Linear Assignment Problem linker (maximum distance = 16 µm, gap-closing ≤ 3 frames). Division events, often missed by classical linkers, were imposed post-hoc with a custom TrackMate-Oneat plug-in. Here, a DenseNet-based “Oneat” (more details in Oneat section) classifier identified mitotic nuclei in cropped 3-D time windows; the plug-in inserted branching points at predicted mitoses and associated daughter tracks within a 16.5 µm search radius, optionally constrained by the MARI (Mitosis Angular Region of Interest) rule to eliminate implausible angular configurations. To use Oneat mitosis detector, aftedr this step we need to perform non-maximal supression to avoid detecting the same event in succeding time frames, you can either do it via non-maximal supression auto script or you can launch our non-maximal supression Napari plugin and also see the mitosis counts prior to and after doing the non-maximal supression.