🧠 Action Classification using Oneat#
🚀 Architecture Overview#
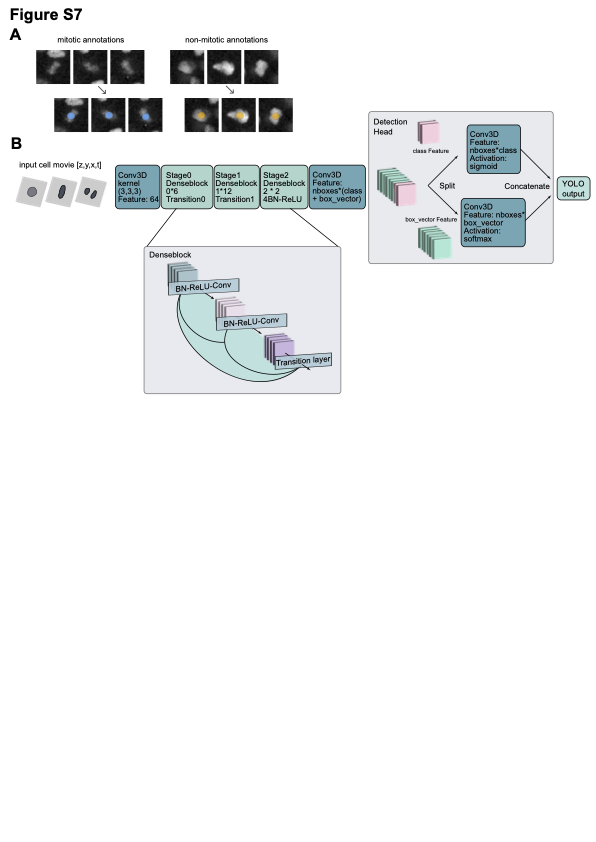
Oneat’s core network, DenseVollNet, processes short 4D image crops by treating time frames as input channels and applying only spatial convolutions (Z, Y, X):
Input:
Patches of size(Z, Y, X)with T timepoints folded as C = T channels (shape(Z, Y, X, T)).DenseVollNet Backbone:
Initial 3D Conv
Conv3Dwithstartfilterfilters, kernel(k_z, k_y, k_x)(e.g.7×7×7),padding='same'BatchNorm → ReLU
Three Dense Block Stages
Each stage i has
depth_ilayers:Bottleneck:
Conv3D(1×1×1), reducing channels (4·F)Feature:
Conv3D(mid_kernel, mid_kernel, mid_kernel), growth rateFConcat the new features with previous tensor
Transition layers between stages (except after the last):
Conv3D(1×1×1)to compress channels (reductionfactor)MaxPool3D(2×2×2)to downsample spatial dimsDownsampling factor per pool: 2
Total downsampling factor: 4 (after two pools), e.g.
(8,64,64) → (2,16,16)
BN → ReLU after final dense block
Fully-Convolutional Head:
A single large
Conv3D(kernelmid_kernel³, padding=’valid’) replaces FC layers, outputtingcategories + nboxes·box_vectorchannels.Kernel size =
(Z/4, Y/4, X/4)=(2,16,16)for input(8,64,64)andlast_conv_factor=4.This collapses the spatial map to
1×1×1per channel.
Split these channels into:
Classification map (
categorieschannels) → SoftmaxRegression map (
nboxes·box_vectorchannels) → Sigmoid
Concat classification & regression outputs.
This design lets Oneat scan any (Z, Y, X) volume with T frames in one pass, yielding per-voxel mitosis predictions.
⚙️ Oneat Mitosis Detector#
Input crops:
64×64×8voxels overTtimepoints (folded into channels).Training samples:
Positive: Napari-clicked mitotic events
Negative: Random non-dividing crops
Loss: Binary cross-entropy
After training, Oneat predicts mitosis coordinates (t, z, y, x) in whole 4D stacks, which the TrackMate-Oneat plugin uses to:
Insert trajectory branches at predicted mitoses
Link daughter cells within a 16.5 µm radius (Jaqaman linker)
Optionally apply MARI to enforce perpendicular daughter positioning, reducing false positives
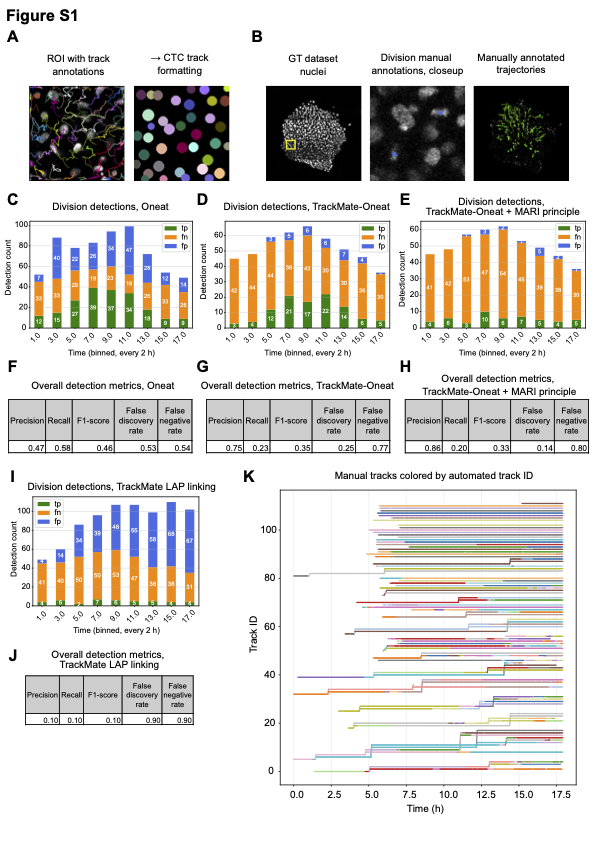
Performance:
Integrating Oneat boosts the precision from 0.1 to 0.86 (with MARI) vs. native TrackMate, with a false discovery rate of 0.14 compared to 0.9 of native TrackMate. TrackMate-Oneat extension uses the same track linking algorithm as TrackMate but with a biologically prior information of the locations of mitotic mother cells and it is only desinged to boost the track linking/Branch Correctness index, the mother cells for which both the daughter cells can not be linked are corrected at a later stage to be classified as a mitotic trajectory.